

Marine fish are known to be rich in docosahexaenoic acid (DHA). However, most marine fish are unable to synthesize DHA themselves. Instead, they ingest and accumulate DHA produced by marine microalgae through the food chain. Therefore, when cultivating marine fish, essential fatty acid requirements are satisfied by adding fish oils containing high levels of DHA to fish feed. It has also been pointed out that fish oil contains various fat-soluble contaminants because the oil is extracted from wild fish that are caught in large quantities. Consequently, there are high expectations for the development of aquaculture technology using vegetable oil, which is known to contain few contaminants. In these marine fish, there is known to be a partial deficit of some of various fatty acid metabolizing enzyme activities such as “Fatty Acid Desaturase 2 (Fads2)” and “Elongation of Very Long chain fatty acids protein (Elovl)” which are necessary for synthesizing DHA from alpha-linolenic acid (ALA). On the other hand, freshwater fish retain Fads2 and Elovl for synthesizing DHA from ALA because DHA is extremely scarce in their food environment. Therefore, my research group sought to improved DHA production capacity by using gene recombination technology to overexpress freshwater fish-derived Fads2 within various fish individuals (Alimmudin et al., 2005; Kabeya et al., 2014) (Figure 1).
 Figure 1: Fatty acid composition of nibea fish liver Figure 1: Fatty acid composition of nibea fish liver
when introducing the fads2 gene of the masu salmon
However, the use of genetically modified fish in aquaculture is unrealistic because the market is adverse to genetic modification technology. Consequently, we focused on genome editing technology that can modify protein function by slightly modifying the endogenous gene sequence. The biggest difficulty in carrying out this experiment is how to modify the amino acid sequence to change Fads2 of marine fish, which cannot synthesize DHA, into a type capable of synthesizing DHA. To overcome this difficulty, we sought hints from a small flatfish that lives in the Amazon River.
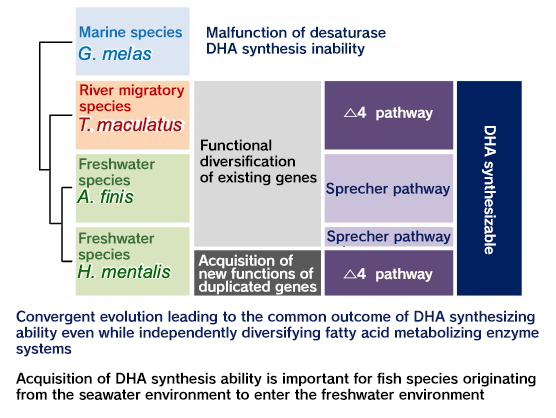 Figure 2: Acquisition style of DHA synthetic ability Figure 2: Acquisition style of DHA synthetic ability
in freshwater flatfish
The ancestral species of flatfish belonging to the Achilles family are distributed only in the sea, but some species have entered into the Amazon River. Upon analyzing the metabolic activity of these fatty acids in the ancestral marine flatfish species, we confirmed that they lack the Δ4 desaturation activity essential for DHA synthesis and the Δ6 desaturation activity for tetracosapentaenoic acid (TPA). However, several species of flatfish in the Achilles family, which are closely related to this species, have adapted to live in freshwater areas that contain almost no DHA in their food environment, and have become able to live entirely in freshwater areas. As a result, we discovered the following examples in freshwater species: (1) Fads2 is gene-duplicated and a newly generated enzyme acquires Δ4 activity, (2) Fads2 is multifunctional and a single enzyme acquires Δ4, 5, and 6 activity, (3) Fads2 is multifunctional and a single enzyme acquires Δ6 activity against TPA in addition to Δ6 activity against ALA. These new enzymatic activities were acquired by slight amino acid mutations in Fads2 of marine flatfish, which are close relatives of these freshwater species (Matsushita et al., 2020) (Fig. 2). Similar tri-functionalization of Fads2 also occurred in freshwater flatfish from Papua New Guinea. This species belongs to a taxonomic group separate from the above-mentioned Achilles family. When considered together with the acquisition of various DHA synthesis strategies of the above-mentioned Achilles family freshwater flatfish, it was strongly suggested that acquisition of DHA-synthesizing ability is essential for fish species of marine origin to advance into freshwater.
Currently, we are working to acquire multi-functionality by referring to such changes in enzyme activity observed in nature and using genome editing to introduce Fads2 amino acid sequence substitutions in marine flatfish. In fact, by substituting only 9 amino acids in the Fads2 sequence derived from Japanese marine flatfish, it is possible to realize tri-functionalization with all enzymatic activities of Δ4, 5, and 6. We were able to confirm this through recombinant protein production systems using yeast hosts. We then performed genome editing by microinjecting the CRISPR/Cas9 system into fertilized eggs. Until now, we have succeeded in establishing parent generations with amino acid substitutions at the target sites. We now aim to mass-produce genome-edited individuals by producing these next generations. These genome-edited individuals are expected to be able to synthesize DHA from ALA on their own. Therefore, it is expected that fish oil-free aquaculture can be realized by adding vegetable oil to feed. If realized, there is hope for the possibility of transforming from the conventional aquaculture industry that consumes DHA into an industry that produces DHA.
Citations
Alimuddin, G. Yoshizaki, V. Kiron, S. Satoh and T. Takeuchi. Enhancement of EPA and DHA biosynthesis by over-expression of masu salmon Δ6-desaturase-like gene in zebrafish. Transgenic Research 14, 159-165. 2005.
Kabeya, N., Y. Takeuchi, Y. Yamamoto, R. Yazawa, Y. Haga, S. Satoh, G. Yoshizaki. Modification of the n-3 HUFA biosynthetic pathway by transgenesis in a marine teleost, nibe croaker. Journal of Biotechnology 172, 46-54. 2014.
Matsushita, Y., K. Miyoshi, N. Kabeya, S. Sanada, R. Yazawa, Y. Haga, S. Satoh, Y. Yamamoto, C. A. Strussmann, A. Luckenbach, G. Yoshizaki. Flatfishes colonised freshwater environments by acquisition of various DHA biosynthetic pathways. Communications Biology 3, 516. 2020.
|